[ad_1]
When a client is identified with cancer, a person of the most essential measures is assessment of the tumor under a microscope by pathologists to decide the cancer phase and to characterize the tumor. This info is central to knowledge medical prognosis (i.e., likely affected person results) and for pinpointing the most acceptable remedy, such as undergoing surgical procedures by itself as opposed to operation moreover chemotherapy. Producing device learning (ML) tools in pathology to help with the microscopic review signifies a persuasive study space with several likely programs.
Preceding scientific studies have shown that ML can accurately detect and classify tumors in pathology visuals and can even forecast client prognosis making use of acknowledged pathology characteristics, these as the diploma to which gland appearances deviate from normal. Though these efforts aim on applying ML to detect or quantify known attributes, different methods offer the probable to recognize novel features. The discovery of new capabilities could in convert more boost cancer prognostication and treatment method decisions for individuals by extracting facts that isn’t nevertheless regarded in existing workflows.
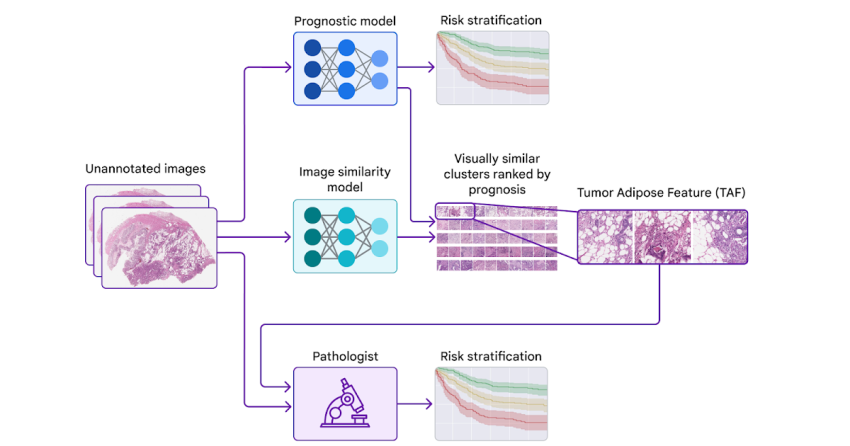
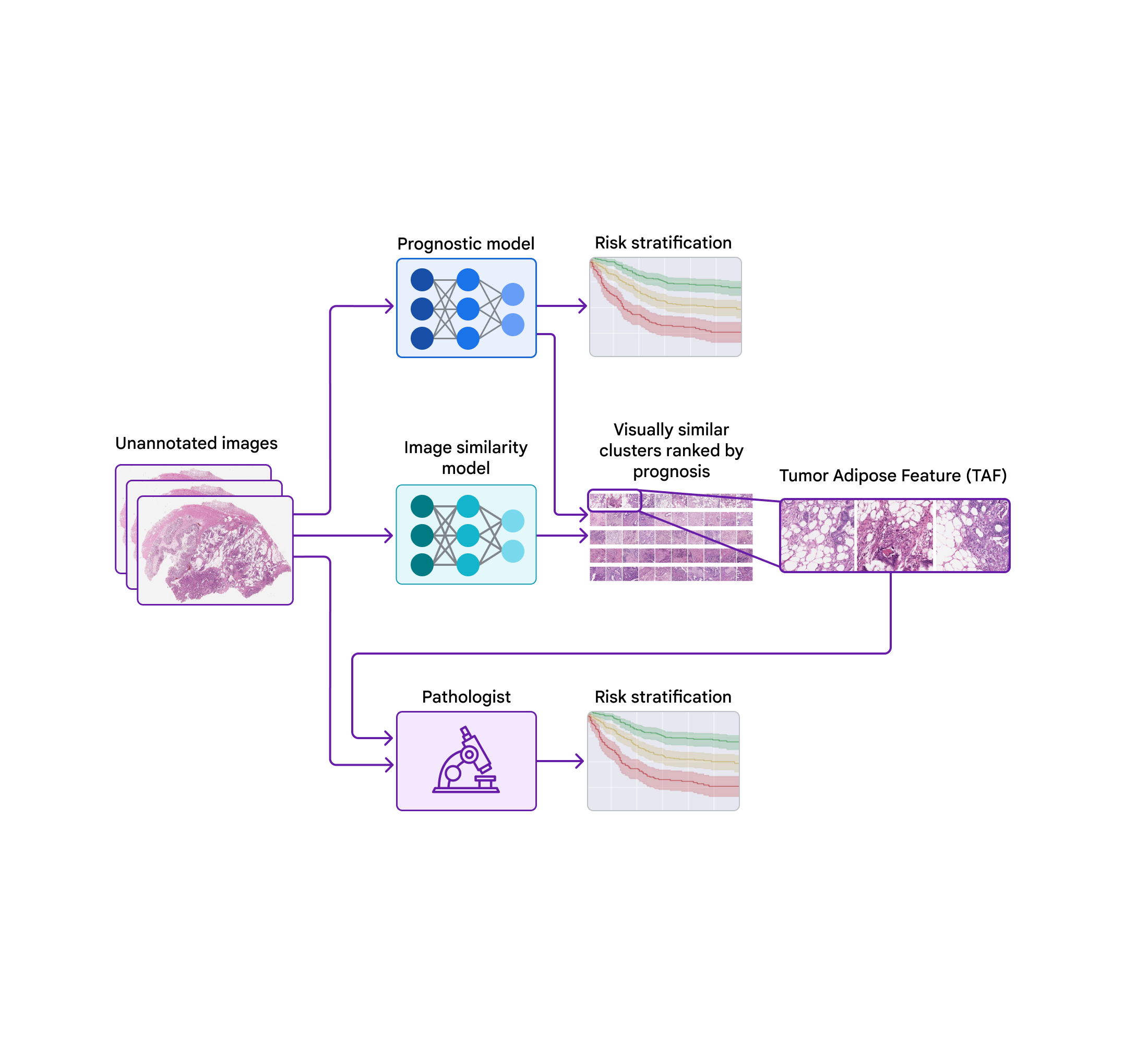
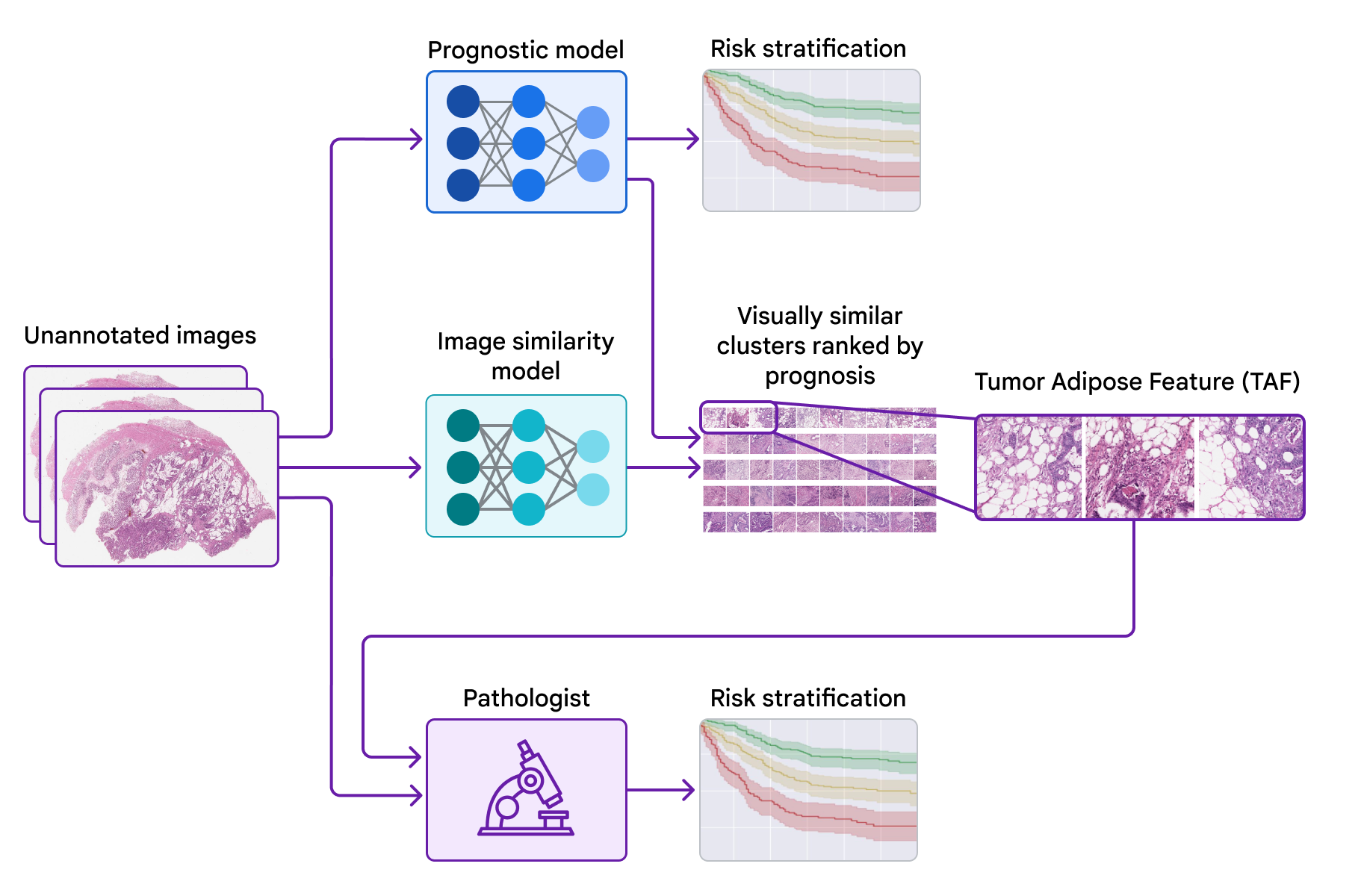
Today, we’d like to share progress we’ve produced above the earlier few years towards figuring out novel functions for colorectal most cancers in collaboration with teams at the Health care University of Graz in Austria and the University of Milano-Bicocca (UNIMIB) in Italy. Underneath, we will protect many phases of the work: (1) coaching a design to forecast prognosis from pathology images devoid of specifying the functions to use, so that it can understand what options are significant (2) probing that prognostic model employing explainability procedures and (3) pinpointing a novel attribute and validating its affiliation with affected person prognosis. We explain this attribute and consider its use by pathologists in our not too long ago posted paper, “Pathologist validation of a device-figured out aspect for colon cancer possibility stratification”. To our knowledge, this is the to start with demonstration that health care authorities can discover new prognostic characteristics from device learning, a promising begin for the long run of this “learning from deep learning” paradigm.
Training a prognostic design to study what functions are critical
One particular potential technique to identifying novel options is to prepare ML styles to directly predict affected individual outcomes utilizing only the photographs and the paired consequence facts. This is in contrast to teaching models to forecast “intermediate” human-annotated labels for recognised pathologic attributes and then working with people features to forecast outcomes.
Initial do the job by our staff showed the feasibility of coaching designs to directly forecast prognosis for a range of most cancers sorts applying the publicly readily available TCGA dataset. It was primarily exciting to see that for some cancer sorts, the model’s predictions were prognostic following controlling for accessible pathologic and scientific features. Together with collaborators from the Medical College of Graz and the Biobank Graz, we subsequently prolonged this perform employing a massive de-recognized colorectal most cancers cohort. Decoding these design predictions grew to become an intriguing up coming action, but prevalent interpretability strategies have been challenging to apply in this context and did not present obvious insights.
Interpreting the model-discovered capabilities
To probe the options applied by the prognostic product, we made use of a second product (qualified to identify graphic similarity) to cluster cropped patches of the huge pathology photos. We then used the prognostic design to compute the regular ML-predicted threat rating for just about every cluster.
Just one cluster stood out for its substantial common threat score (involved with weak prognosis) and its unique visible physical appearance. Pathologists described the visuals as involving higher grade tumor (i.e., the very least-resembling typical tissue) in close proximity to adipose (excess fat) tissue, main us to dub this cluster the “tumor adipose feature” (TAF) see following determine for in depth illustrations of this characteristic. Further examination showed that the relative quantity of TAF was by itself remarkably and independently prognostic.
| Remaining: H&E pathology slide with an overlaid heatmap indicating places of the tumor adipose aspect (TAF). Regions highlighted in crimson/orange are considered to be extra possible TAF by the graphic similarity product, compared to locations highlighted in inexperienced/blue or locations not highlighted at all. Correct: Agent collection of TAF patches across multiple situations. |
Validating that the product-realized characteristic can be utilized by pathologists
These experiments provided a compelling example of the opportunity for ML models to predict affected individual outcomes and a methodological approach for getting insights into product predictions. On the other hand, there remained the intriguing issues of no matter whether pathologists could learn and rating the attribute determined by the design although maintaining demonstrable prognostic benefit.
In our most recent paper, we collaborated with pathologists from the UNIMIB to investigate these concerns. Employing example visuals of TAF from the previous publication to find out and fully grasp this element of desire, UNIMIB pathologists developed scoring rules for TAF. If TAF was not noticed, the scenario was scored as “absent”, and if TAF was noticed, then “unifocal”, “multifocal”, and “widespread” classes ended up made use of to point out the relative amount. Our study confirmed that pathologists could reproducibly discover the ML-derived TAF and that their scoring for TAF provided statistically sizeable prognostic benefit on an unbiased retrospective dataset. To our knowledge, this is the initially demonstration of pathologists learning to discover and score a distinct pathology characteristic originally recognized by an ML-dependent method.
Placing things in context: discovering from deep studying as a paradigm
Our get the job done is an example of people today “learning from deep learning”. In common ML, designs study from hand-engineered functions educated by current domain know-how. A lot more a short while ago, in the deep studying era, a mix of substantial-scale design architectures, compute, and datasets has enabled learning straight from uncooked information, but this is normally at the cost of human interpretability. Our function couples the use of deep understanding to forecast affected individual outcomes with interpretability approaches, to extract new know-how that could be applied by pathologists. We see this procedure as a purely natural up coming phase in the evolution of applying ML to troubles in drugs and science, moving from the use of ML to distill current human information to men and women utilizing ML as a device for expertise discovery.
Acknowledgements
This get the job done would not have been attainable without the attempts of coauthors Vincenzo L’Imperio, Markus Plass, Heimo Muller, Nicolò’ Tamini, Luca Gianotti, Nicola Zucchini, Robert Reihs, Greg S. Corrado, Dale R. Webster, Lily H. Peng, Po-Hsuan Cameron Chen, Marialuisa Lavitrano, David F. Steiner, Kurt Zatloukal, Fabio Pagni. We also recognize the guidance from Verily Life Sciences and the Google Overall health Pathology teams – in unique Timo Kohlberger, Yunnan Cai, Hongwu Wang, Kunal Nagpal, Craig Mermel, Trissia Brown, Isabelle Flament-Auvigne, and Angela Lin. We also enjoy manuscript suggestions from Akinori Mitani, Rory Sayres, and Michael Howell, and illustration aid from Abi Jones. This operate would also not have been feasible without having the help of Christian Guelly, Andreas Holzinger, Robert Reihs, Farah Nader, the Biobank Graz, the efforts of the slide digitization group at the Healthcare University Graz, the participation of the pathologists who reviewed and annotated circumstances through design growth, and the professionals of the UNIMIB workforce.
[ad_2]
Source connection