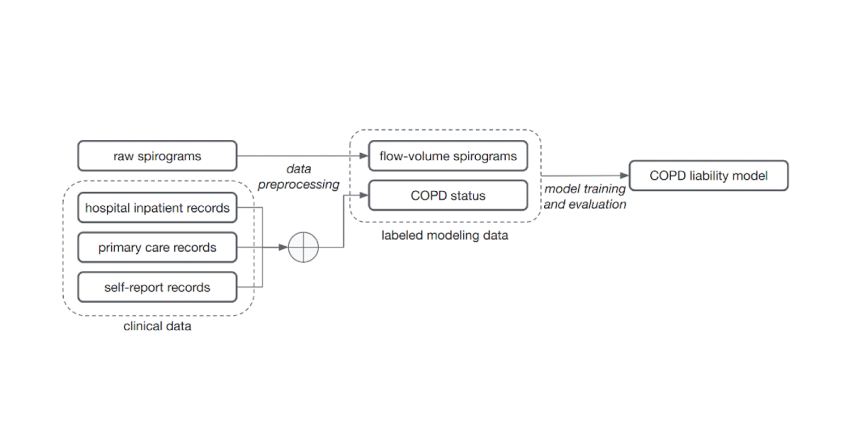
[ad_1]
The mix of the atmosphere an particular person encounters and their genetic predispositions determines the the vast majority of their hazard for many diseases. Significant national endeavours, this sort of as the United kingdom Biobank, have created substantial, general public resources to greater fully grasp the backlinks involving natural environment, genetics, and ailment. This has the opportunity to enable men and women much better understand how to continue to be healthier, clinicians to handle ailments, and experts to create new medicines.
A person problem in this course of action is how we make perception of the wide amount of medical measurements — the United kingdom Biobank has many petabytes of imaging, metabolic exams, and medical documents spanning 500,000 persons. To very best use this data, we will need to be equipped to depict the details existing as succinct, useful labels about meaningful illnesses and characteristics, a procedure called phenotyping. That is in which we can use the ability of ML designs to select up on delicate intricate designs in big quantities of info.
We have formerly demonstrated the ability to use ML styles to quickly phenotype at scale for retinal ailments. Nonetheless, these types were skilled applying labels from clinician judgment, and entry to clinical-grade labels is a restricting variable thanks to the time and expense needed to build them.
In “Inference of serious obstructive pulmonary disorder with deep studying on raw spirograms identifies new genetic loci and increases danger styles”, released in Character Genetics, we’re psyched to emphasize a method for instruction accurate ML designs for genetic discovery of conditions, even when employing noisy and unreliable labels. We reveal the skill to practice ML types that can phenotype straight from uncooked medical measurement and unreliable medical report information. This minimized reliance on health care domain authorities for labeling drastically expands the variety of applications for our system to a panoply of health conditions and has the prospective to enhance their prevention, diagnosis, and cure. We showcase this process with ML styles that can greater characterize lung functionality and continual obstructive pulmonary disease (COPD). Furthermore, we exhibit the usefulness of these styles by demonstrating a far better capability to establish genetic variants involved with COPD, enhanced knowledge of the biology behind the sickness, and effective prediction of results associated with COPD.
ML for deeper understanding of exhalation
For this demonstration, we focused on COPD, the third major result in of around the globe loss of life in 2019, in which airway inflammation and impeded airflow can progressively minimize lung function. Lung functionality for COPD and other illnesses is calculated by recording an individual’s exhalation volume about time (the document is known as a spirogram see an case in point under). Whilst there are pointers (called GOLD) for pinpointing COPD status from exhalation, these use only a couple of, distinct knowledge factors in the curve and apply fastened thresholds to people values. Much of the loaded knowledge from these spirograms is discarded in this evaluation of lung function.
We reasoned that ML styles experienced to classify spirograms would be able to use the wealthy details present additional fully and consequence in extra accurate and in depth steps of lung perform and disorder, related to what we have seen in other classification responsibilities like mammography or histology. We properly trained ML products to predict whether an personal has COPD making use of the full spirograms as inputs.
The popular approach of teaching styles for this trouble, supervised finding out, calls for samples to be related with labels. Determining these labels can need the energy of really time-constrained industry experts. For this function, to clearly show that we do not always will need medically graded labels, we decided to use a selection of broadly offered resources of health-related report information and facts to produce these labels with out healthcare specialist evaluation. These labels are a lot less trusted and noisy for two reasons. Initially, there are gaps in the clinical documents of persons because they use many well being solutions. Second, COPD is generally undiagnosed, indicating many with the illness will not be labeled as having it even if we compile the comprehensive clinical information. Nonetheless, we skilled a product to predict these noisy labels from the spirogram curves and deal with the product predictions as a quantitative COPD legal responsibility or risk score.
Predicting COPD outcomes
We then investigated whether the chance scores developed by our model could superior predict a variety of binary COPD results (for instance, an individual’s COPD status, regardless of whether they ended up hospitalized for COPD or died from it). For comparison, we benchmarked the model relative to pro-outlined measurements expected to diagnose COPD, particularly FEV1/FVC, which compares certain points on the spirogram curve with a simple mathematical ratio. We observed an advancement in the skill to predict these results as noticed in the precision-remember curves under.
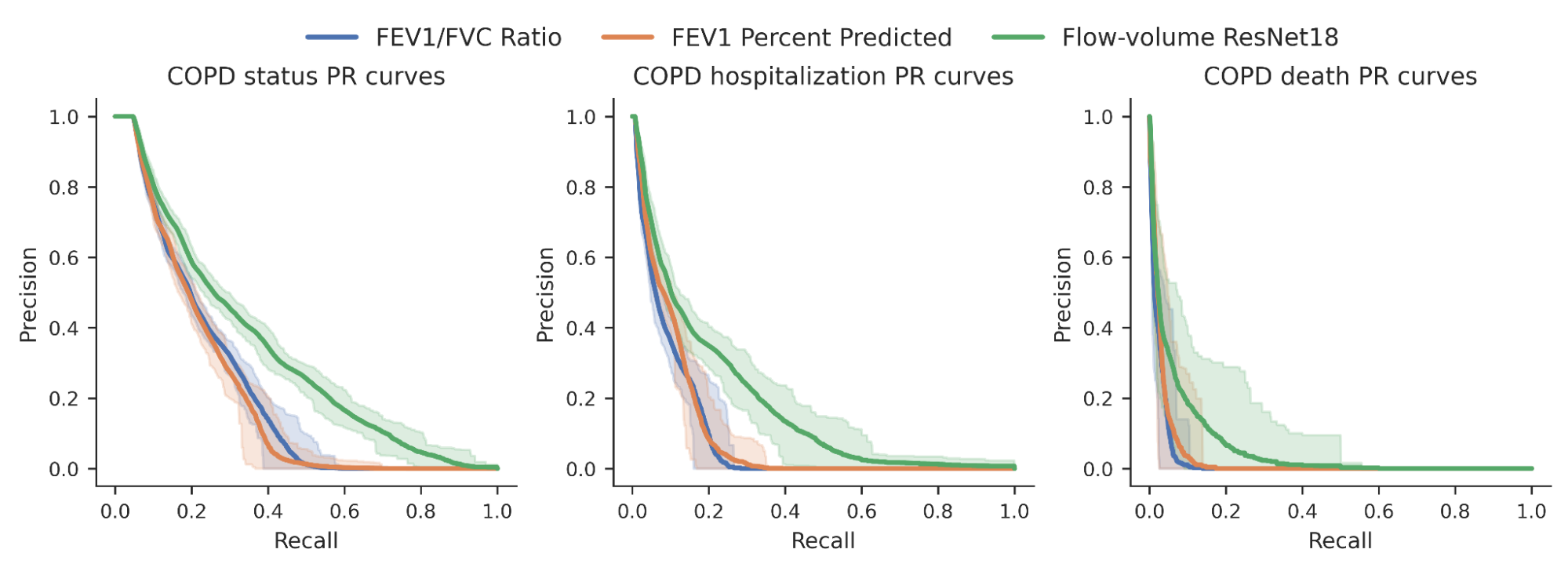 |
| Precision-recall curves for COPD standing and results for our ML model (eco-friendly) in contrast to standard steps. Assurance intervals are proven by lighter shading. |
We also observed that separating populations by their COPD product rating was predictive of all-result in mortality. This plot indicates that individuals with bigger COPD chance are extra probable to die earlier from any brings about and the chance probably has implications over and above just COPD.
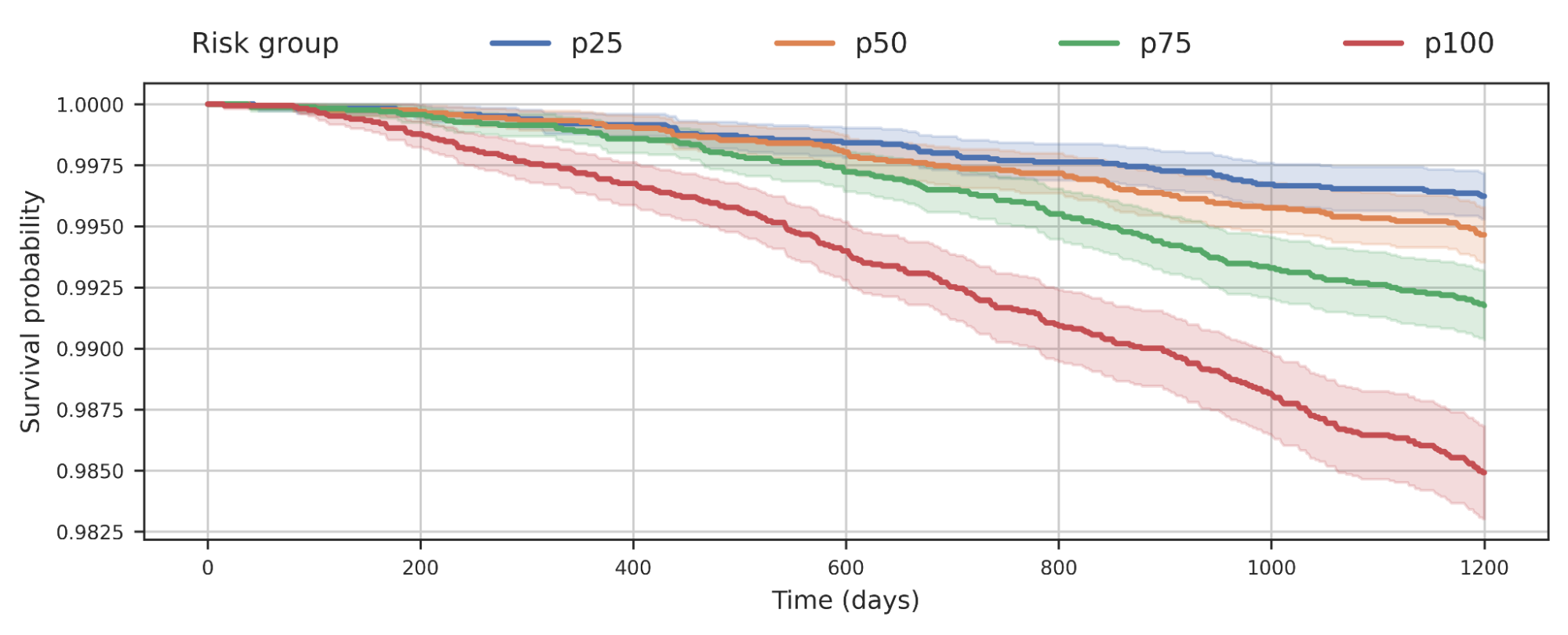 |
| Survival investigation of a cohort of United kingdom Biobank men and women stratified by their COPD model’s predicted hazard quartile. The decrease of the curve suggests people in the cohort dying around time. For illustration, p100 represents the 25% of the cohort with biggest predicted hazard, when p50 represents the 2nd quartile. |
Figuring out the genetic hyperlinks with COPD
Given that the goal of significant scale biobanks is to deliver alongside one another large quantities of both phenotype and genetic data, we also carried out a test termed a genome-large association review (GWAS) to detect the genetic links with COPD and genetic predisposition. A GWAS measures the strength of the statistical association concerning a given genetic variant — a change in a unique place of DNA — and the observations (e.g., COPD) throughout a cohort of conditions and controls. Genetic associations learned in this way can inform drug development that modifies the activity or products and solutions of a gene, as very well as increase our being familiar with of the biology for a sickness.
We showed with our ML-phenotyping method that not only do we rediscover nearly all recognised COPD variants uncovered by manual phenotyping, but we also uncover several novel genetic variants drastically affiliated with COPD. In addition, we see good arrangement on the effect dimensions for the variants discovered by both of those our ML tactic and the guide just one (R2=.93), which gives powerful evidence for validity of the freshly uncovered variants.
Finally, our collaborators at Harvard Clinical College and Brigham and Women’s Medical center more examined the plausibility of these conclusions by supplying insights into the doable biological position of the novel variants in advancement and development of COPD (you can see extra discussion on these insights in the paper).
Conclusion
We demonstrated that our previously procedures for phenotyping with ML can be expanded to a huge assortment of diseases and can provide novel and precious insights. We made two essential observations by utilizing this to forecast COPD from spirograms and exploring new genetic insights. Initially, domain information was not vital to make predictions from raw clinical facts. Curiously, we confirmed the raw medical details is almost certainly underutilized and the ML design can come across styles in it that are not captured by professional-described measurements. 2nd, we do not will need medically graded labels in its place, noisy labels outlined from extensively obtainable professional medical documents can be employed to produce clinically predictive and genetically informative hazard scores. We hope that this do the job will broadly develop the potential of the industry to use noisy labels and will improve our collective comprehension of lung purpose and ailment.
Acknowledgments
This work is the combined output of numerous contributors and establishments. We thank all contributors: Justin Cosentino, Babak Alipanahi, Zachary R. McCaw, Cory Y. McLean, Farhad Hormozdiari (Google), Davin Hill (Northeastern University), Tae-Hwi Schwantes-An and Dongbing Lai (Indiana College), Brian D. Hobbs and Michael H. Cho (Brigham and Women’s Healthcare facility, and Harvard Health care College). We also thank Ted Yun and Nick Furlotte for reviewing the manuscript, Greg Corrado and Shravya Shetty for assist, and Howard Yang, Kavita Kulkarni, and Tammi Huynh for encouraging with publication logistics.
[ad_2]
Source hyperlink