[ad_1]
Getting old is a process that is characterised by physiological and molecular improvements that boost an individual’s chance of building health conditions and inevitably dying. Currently being capable to measure and estimate the biological signatures of aging can help scientists detect preventive measures to reduce illness risk and effects. Researchers have created “aging clocks” centered on markers this kind of as blood proteins or DNA methylation to measure individuals’ organic age, which is unique from one’s chronological age. These getting old clocks enable predict the risk of age-associated health conditions. But mainly because protein and methylation markers need a blood draw, non-invasive approaches to obtain identical actions could make getting older data far more obtainable.
Potentially amazingly, the options on our retinas mirror a great deal about us. Photos of the retina, which has vascular connections to the mind, are a worthwhile supply of organic and physiological data. Its capabilities have been joined to several growing older-connected illnesses, including diabetic retinopathy, cardiovascular sickness, and Alzheimer’s sickness. Also, former do the job from Google has demonstrated that retinal photographs can be made use of to predict age, threat of cardiovascular disorder, or even intercourse or smoking cigarettes status. Could we extend these findings to getting old, and possibly in the process recognize a new, useful biomarker for human disease?
In a new paper “Longitudinal fundus imaging and its genome-vast affiliation evaluation present proof for a human retinal ageing clock”, we exhibit that deep learning types can precisely forecast organic age from a retinal picture and reveal insights that better forecast age-relevant illness in folks. We discuss how the model’s insights can strengthen our comprehending of how genetic components influence getting old. On top of that, we’re releasing the code modifications for these styles, which make on ML frameworks for examining retina photos that we have formerly publicly produced.
Predicting chronological age from retinal images
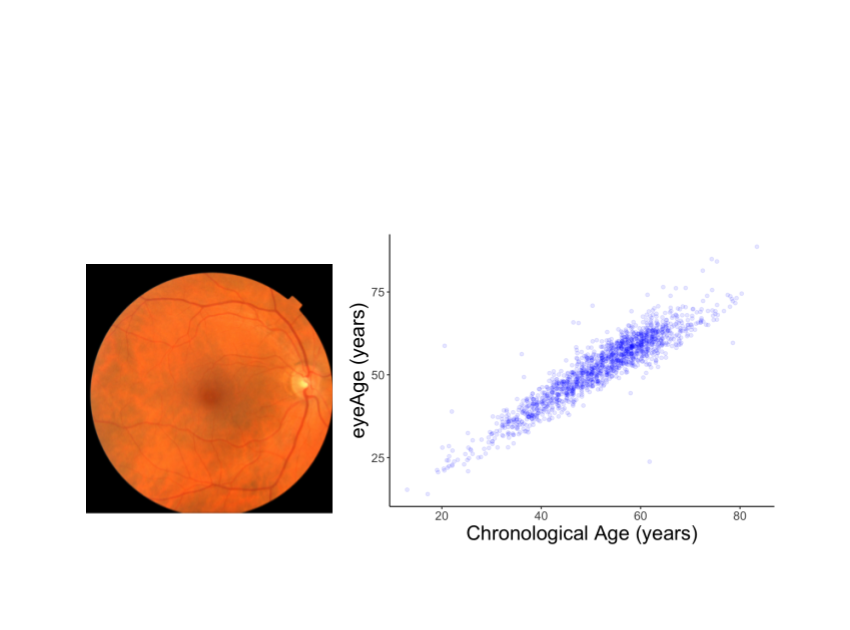
We skilled a design to predict chronological age utilizing hundreds of 1000’s of retinal illustrations or photos from a telemedicine-dependent blindness avoidance plan that were captured in main care clinics and de-recognized. A subset of these photos has been made use of in a opposition by Kaggle and educational publications, like prior Google function with diabetic retinopathy.
We evaluated the resulting design efficiency both equally on a held-out set of 50,000 retinal images and on a individual UKBiobank dataset made up of close to 120,000 pictures. The model predictions, named eyeAge, strongly correspond with the genuine chronological age of men and women (proven underneath Pearson correlation coefficient of .87). This is the to start with time that retinal illustrations or photos have been applied to produce these types of an correct aging clock.
Analyzing the predicted and true age gap
Even however eyeAge correlates with chronological age properly across several samples, the determine over also demonstrates men and women for which the eyeAge differs significantly from chronological age, equally in instances the place the product predicts a worth a lot younger or older than the chronological age. This could indicate that the model is discovering elements in the retinal images that replicate real biological outcomes that are pertinent to the illnesses that grow to be more prevalent with organic age.
To take a look at regardless of whether this change displays fundamental biological factors, we explored its correlation with situations this sort of as continual obstructive pulmonary ailment (COPD) and myocardial infarction and other biomarkers of health like systolic blood strain. We noticed that a predicted age larger than the chronological age, correlates with disease and biomarkers of wellness in these circumstances. For illustration, we confirmed a statistically major (p=.0028) correlation involving eyeAge and all-trigger mortality — that is a higher eyeAge was connected with a greater probability of demise through the study.
Revealing genetic aspects for aging
To even more investigate the utility of the eyeAge product for generating biological insights, we connected product predictions to genetic variants, which are available for men and women in the massive UKBiobank examine. Importantly, an individual’s germline genetics (the variants inherited from your mom and dad) are fastened at birth, making this measure independent of age. This assessment created a listing of genes related with accelerated biological growing older (labeled in the figure below). The prime discovered gene from our genome-huge affiliation analyze is ALKAL2, and apparently the corresponding gene in fruit flies had beforehand been revealed to be included in extending daily life span in flies. Our collaborator, Professor Pankaj Kapahi from the Buck Institute for Study on Growing older, discovered in laboratory experiments that cutting down the expression of the gene in flies resulted in improved eyesight, providing an indication of ALKAL2 influence on the growing old of the visible procedure.
 |
| Manhattan plot representing substantial genes connected with hole concerning chronological age and eyeAge. Significant genes displayed as details over the dotted threshold line. |
Programs
Our eyeAge clock has several likely apps. As demonstrated over, it enables scientists to discover markers for aging and age-related health conditions and to discover genes whose features may be altered by medications to boost healthier growing old. It may perhaps also assistance researchers more recognize the results of life style practices and interventions this sort of as workout, eating plan, and medication on an individual’s biological getting older. On top of that, the eyeAge clock could be beneficial in the pharmaceutical market for evaluating rejuvenation and anti-aging therapies. By tracking changes in the retina around time, scientists may possibly be in a position to ascertain the usefulness of these interventions in slowing or reversing the ageing approach.
Our strategy to use retinal imaging for tracking biological age involves amassing images at numerous time points and analyzing them longitudinally to properly predict the way of getting old. Importantly, this technique is non-invasive and does not need specialized lab devices. Our conclusions also indicate that the eyeAge clock, which is centered on retinal pictures, is unbiased from blood-biomarker–based growing old clocks. This allows researchers to review growing old via a further angle, and when put together with other markers, provides a more comprehensive comprehending of an individual’s organic age. Also as opposed to existing growing older clocks, the considerably less invasive character of imaging (when compared to blood exams) could possibly enable eyeAge to be used for actionable biological and behavioral interventions.
Conclusion
We clearly show that deep discovering types can accurately predict an individual’s chronological age applying only pictures of their retina. In addition, when the predicted age differs from chronological age, this big difference can recognize accelerated onset of age-linked illness. Ultimately, we clearly show that the designs master insights which can strengthen our knowing of how genetic elements influence getting older.
We’ve publicly released the code modifications utilised for these versions which construct on ML frameworks for examining retina photos that we have beforehand publicly unveiled.
It is our hope that this do the job will aid scientists generate greater procedures to establish ailment and sickness threat early, and lead to a lot more helpful drug and life-style interventions to promote nutritious ageing.
Acknowledgments
This do the job is the outcome of the merged initiatives of several teams. We thank all contributors: Sara Ahadi, Boris Babenko, Cory McLean, Drew Bryant, Orion Pritchard, Avinash Varadarajan, Marc Berndl and Ali Bashir (Google Investigate), Kenneth Wilson, Enrique Carrera and Pankaj Kapahi (Buck Institute of Getting old Research), and Ricardo Lamy and Jay Stewart (College of California, San Francisco). We would also like to thank Michelle Dimon and John Platt for reviewing the manuscript, and Preeti Singh for aiding with publication logistics.
[ad_2]
Resource connection